Among the more obscure papers proposing ivermectin as a treatment for Covid-19 there are some that suggest it acts directly by binding to the SARS-CoV-2 spike protein (and other proteins of that virus too), based on in silico (computational) studies:
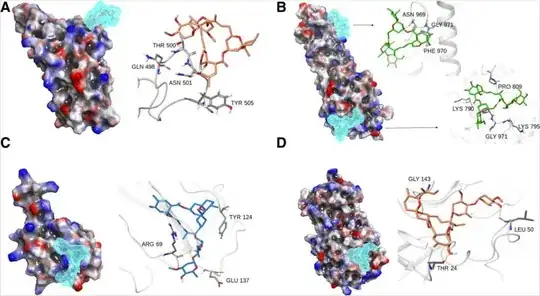
It can be deduced that Ivermectin has efficient binding with: (1) spike S1-RBD, where it binds with Thr 500, Asn 501 and Tyr 505 residues. These sites are critical to the SARS CoV-2 spike protein-mediated recognition from ACE-2 receptors in the host cellular system. Prominent H-bonding with Thr 500 and Asn 501 and water bridges were observed for more than 80% of simulation (Fig. 4a). (2) Spike S2-fusion domain, it binds to two specific regions of S2 fusion domain namely HR-1 sub-domain and fusion peptide domain. Major interactions were observed at the HR-1 domain, where it binds for up to 80% of simulation duration with Ser 968, Asn 969 and Gly 971 with H-bonds and water bridges. The fusion peptide region also exhibits weak affinity for ivermectin at Phe 797 and extremely weak interactions at Pro 792 residues with hydrophobic contact (Figs. 4b). The S2 fusion domain is necessary to build the fusion bridge between the viral and host membrane, where the fusion peptide is highly non-polar flexible region which facilitates the direct contact with the host membrane components. (3) N-protein, the poly-nucleotide (RNA) interacting cleft of nucleocapsid N-protein characterized by residues Arg 69, Tyr 124, Asn 127 and Glu 137 were found interacting with ivermectin with rich H-bond ratio, see Fig 4c. (4) Main protease, the main protease of the SARS-CoV 2 is another target which exhibits good affinity for ivermectin in inhibition site too at Glu 19, Thr 25, Glu 47, Leu 50 (Fig. 4d).
On the other hand, the somewhat more cited papers on ivermectin don't mention that (putative) direct mechanism of action on SARS-CoV-2 at all and only mention host-directed (IMPα-directed) effects via as the plausible mechanism. (Effects of this latter kind would not be specific to particular viruses, but result in broad anti-viral action, as I understand the issue.)
So are there in vitro (as opposed to in silico) studies that find any direct effect (i.e. affinity/avidity) of ivermectin to SARS-CoV-2 proteins as suggested in that first study? (Such lab studies are normally conducted with pseudoviral particles with just the protein of interest, at least for the spike protein; I'm not sure how one could test the main protease (aka 3CLpro) in that regard though.)